Summary
The reappearance and settlement of the grey wolf in eastern and northern Germany over the last 15 years has been a focal point of environmental discourse. Allowing passages of wilderness to bifurcate the land bolsters dwindling population numbers but increases the chances of contact with settlements and habitations.
Here I retrieved data to gain some personal insight into the spatial and temporal expansion of wolves over the last +/- 15 years.
Data Preparation
# load libraries
library(readr)
library(tidyverse)
library(DT)
library(deldir)
# load custom functions
source("static/data/ROAM.r")# load locale data
wolf_data <- read_delim("static/data/wolves/wolf_data_2014.tsv",
"\t", escape_double = FALSE, trim_ws = TRUE)One can include the implicit NAs for each year-location pair, and fill missing observations to build a “complete dataset”.
# complete the data by filling missing "data-slots"
complete.ts <-wolf_data %>%
complete(Jahr=full_seq(Jahr,1),nesting(Bundesland,Territorium),
fill=list(Status="np",Repro="na",Welpen=0))
# make searchable table of complete data
complete.ts %>%
datatable(., rownames = FALSE, filter="top",
options = list(pageLength = 10, scrollX=F)) %>%
DT::formatStyle(columns = c(1:6), fontSize = '85%')One can create a lookup-table to hold the human-readable labels of the factors in the data.
# create annotations
labs_status <-data.frame(Status=c("e","p","r","np"),
Name=c("Single","Pair","Pack","Not Present"),
stringsAsFactors = FALSE)Aggregating the Temporal Data
The wolves have aggregated in 2 main locations (as of 2014), namely Noctum and Neustadt.
# find unique locations with newborns
locations <- complete.ts %>%
group_by(Territorium) %>%
summarise(welpen_sum = sum(Welpen)) %>%
arrange(desc(welpen_sum)) %>% filter(.,welpen_sum>0)
# most prevalent breeding locations
locations_select <- complete.ts %>%
group_by(Territorium) %>%
summarise(welpen_sum = sum(Welpen)) %>%
arrange(desc(welpen_sum)) %>% filter(.,welpen_sum>10)
# quick plot
locations_select %>%
ggplot(.,aes(fct_reorder(Territorium,welpen_sum,.desc = TRUE),welpen_sum)) +
geom_col() +
theme_plain(base_size = 10) +
scale_x_discrete(labels= abbreviate) +
xlab("Territory") + ylab("Total Newborns (n)") +
labs(title="Aggregated Count of Wolves in Eastern Germany, 2001-2014")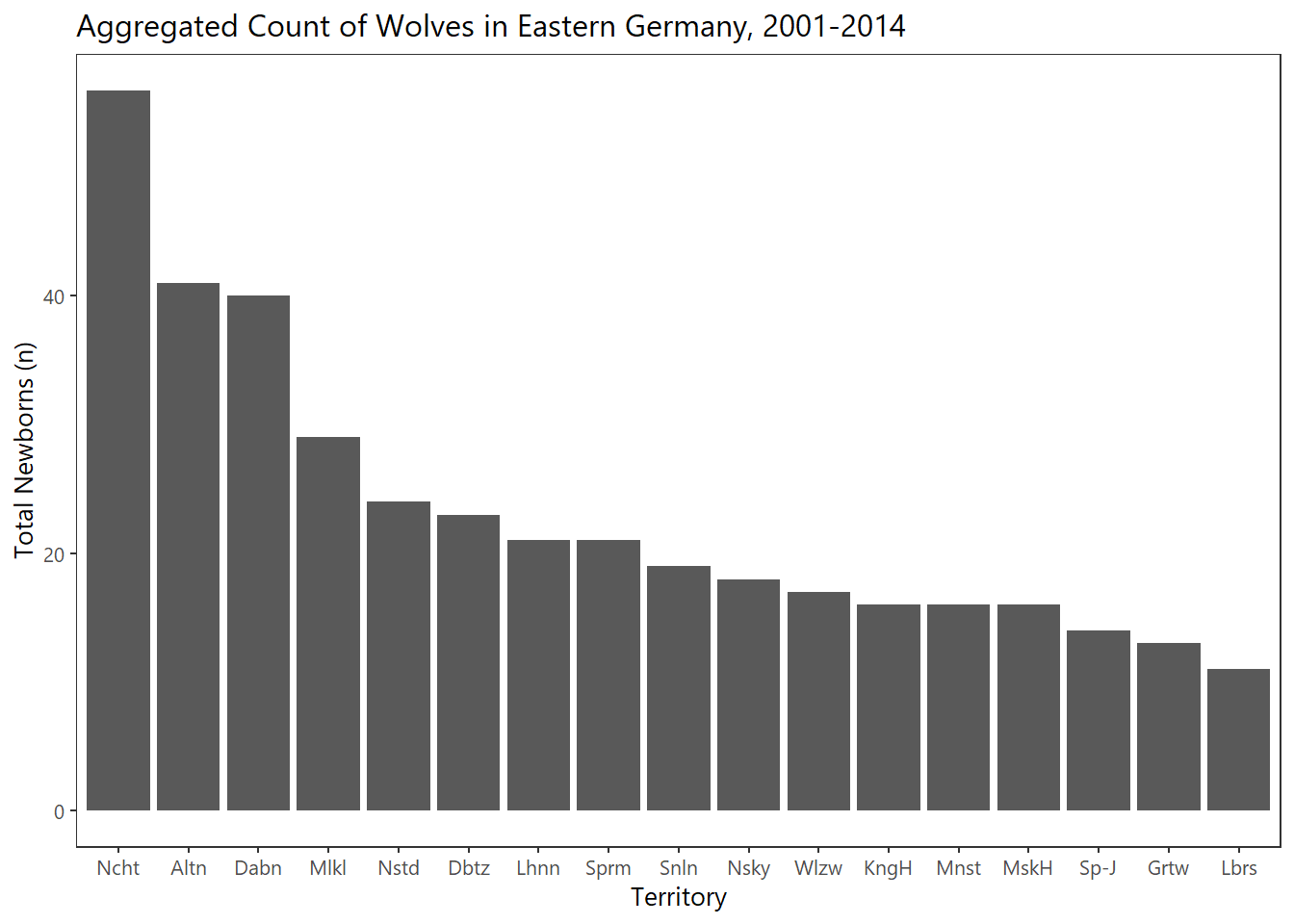
Spatial Distribution as a Proxy for Interaction Encounters
Spatial information (Lat/Lon) was manually retrieved using the www.latlon.net service.
# load coordinates information
coords_wolves <- read_delim("ststic/data/wolves/coords_wolves_2014.tsv","\t",
escape_double = FALSE, trim_ws = TRUE)
# join with locale data
locations_mapped <-left_join(locations,coords_wolves)One can quickly build up a figure which depicts the:
- aggregated total of newborns (+existing wolves) at any location
- draw this using an appropriate map projection (in this case AZ Equal Area)
- add the tessselation segments
- compute a 2d kernel density of the spatial distribution
A Voronoi Tesselation splits a plane into segments given by points ditributed within it so that each segment contains the point and the area which is closer to that point that any other
# compute voronoi tesselation
voronoi_gg <- deldir(locations_mapped$Lon,locations_mapped$Lat)
# build voronoi and density plot
ggplot(data=locations_mapped,aes(Lon,Lat)) +
geom_segment(aes(x=x1,y=y1,xend=x2,yend=y2),
size=0.5,color="grey",data=voronoi_gg$dirsgs) +
stat_density_2d(aes(fill = ..level..),
geom = "polygon", alpha = 0.25, color = NA) +
scale_fill_gradient2("Density", low = "white", mid = "yellow", high = "red", midpoint = 0.25) +
geom_point(size=log(locations_mapped$welpen_sum)) +
xlab("Longitude (degrees east)") + ylab("Latitude (degrees north)") +
labs(title="Wolf Prospensity in Eastern Germany",
subtitle="With Voronoi Tesselations Marking Locales (AZ Equal Area Projection)") +
theme_plain(base_size = 11) +
coord_map("azequalarea",orientation = c(51.5,13,5)) +
scale_y_continuous(limits=c(50,53),
breaks=c(50,51.5,53)) +
scale_x_continuous(limits=c(8,16),
breaks=c(8,10,12,14,16))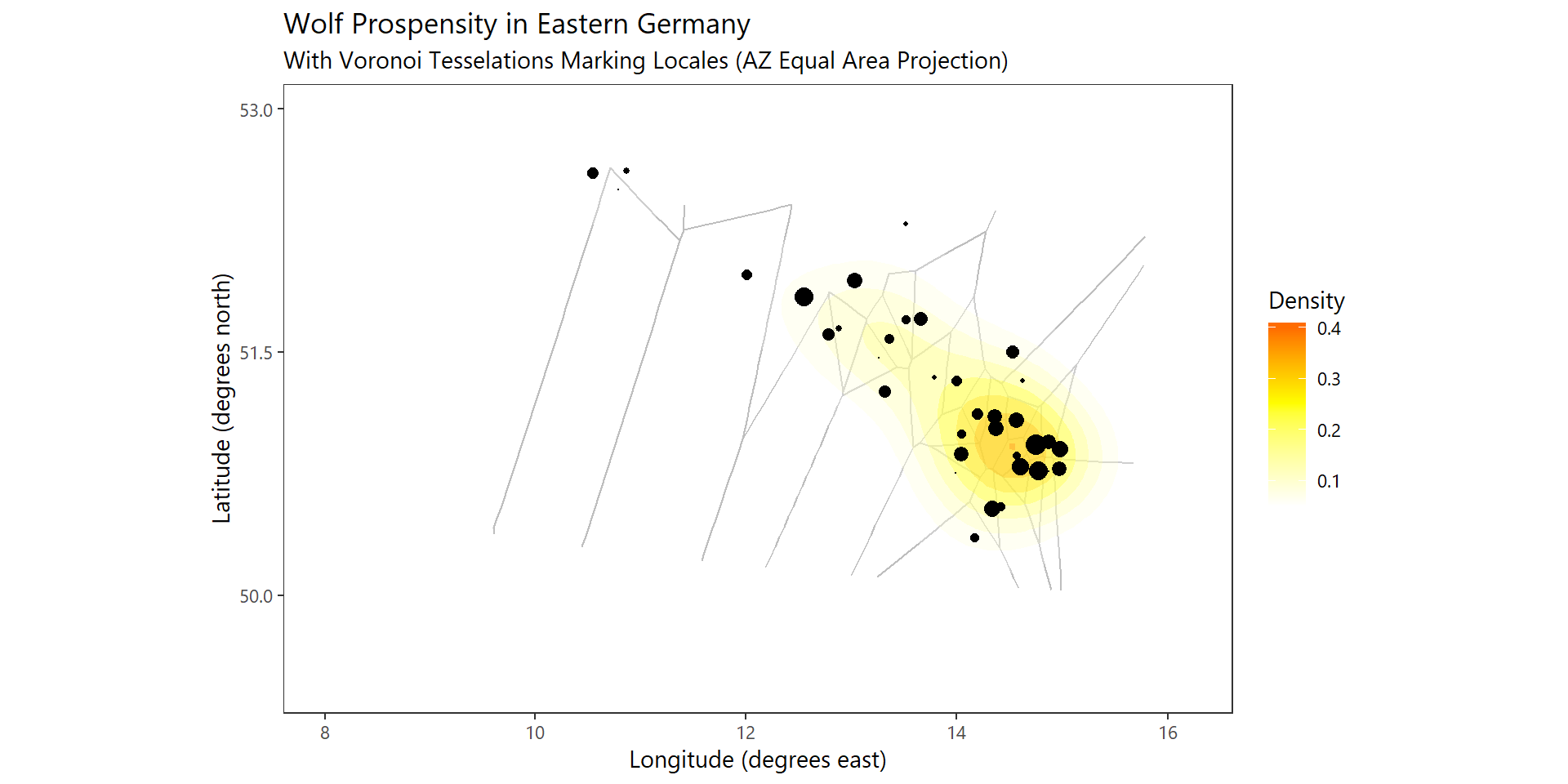
Plot an overview dashboard of population growth over time, in selected regions.
# select groups of interest
target_locales <- c("Sachsen", "Sachsen-Anhalt","Brandenburg", "Niedersachsen")
target_groups <- c("r","p")
stats <- complete.ts %>%
group_by(Jahr,Bundesland,Status) %>%
summarise(n=n()) %>% filter(., Status %in% target_groups) %>%
filter(.,Bundesland %in% target_locales) %>%
left_join(labs_status)
# plot :: ts of groups over time per region
ggplot(stats,aes(Jahr,n)) + geom_point() +
geom_smooth(method="loess",se=FALSE) +
facet_grid(Bundesland ~ Name) +
theme_plain(base_size = 11) +
xlab("Year") + ylab("Count") +
scale_y_continuous(expand=c(0.1,0.1),
breaks=c(0,5,10)) +
scale_x_continuous(expand=c(0.1,0.1),
breaks=c(2000,2004,2008,2012,2016))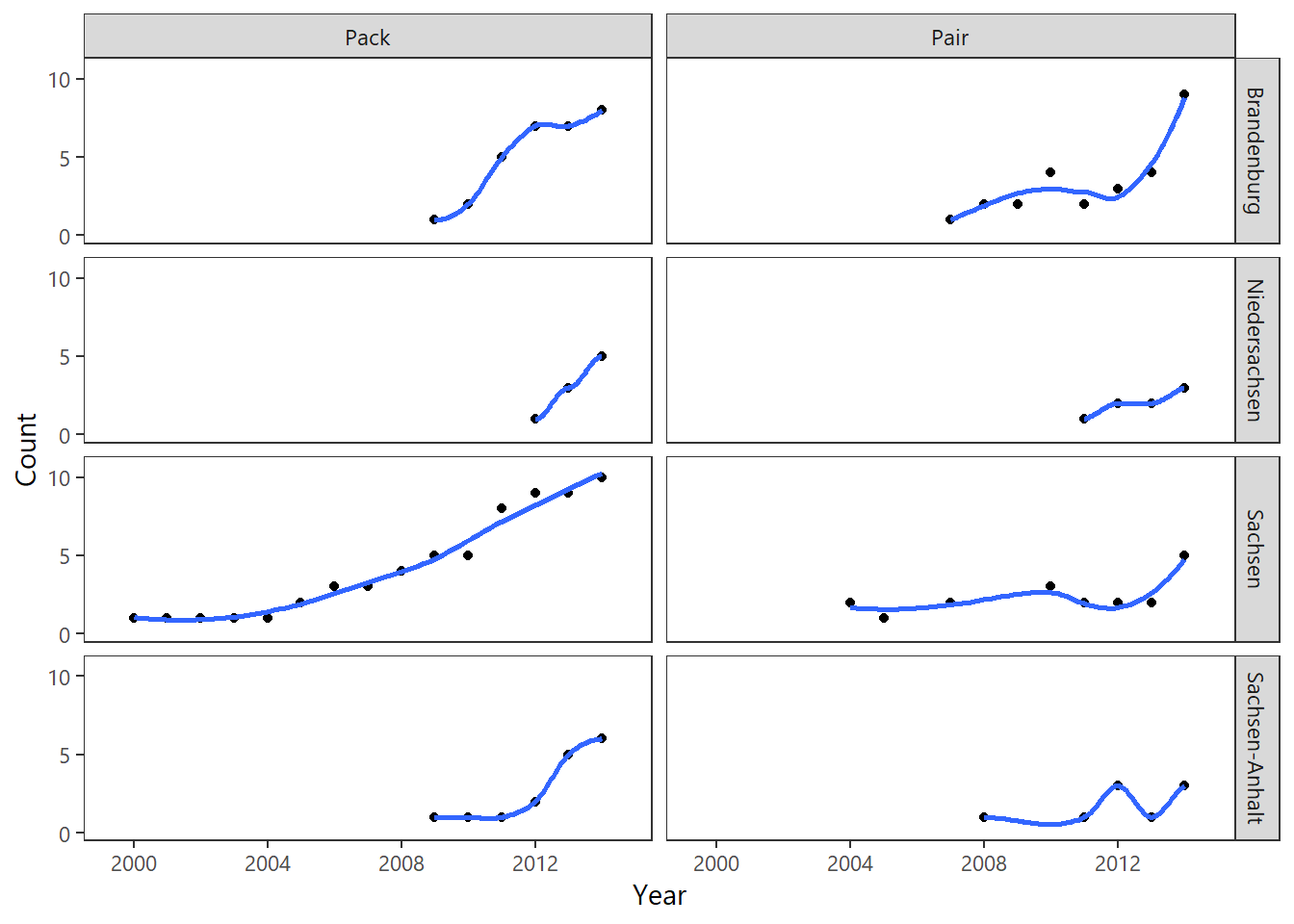
Build a complete map, with informative map layers, extra rasters and groupings.
library(leaflet)
library(leaflet.extras)
# work in progress :: testing heatmap functionality
leaflet(data = locations_mapped) %>%
addProviderTiles(providers$Esri.NatGeoWorldMap) %>%
setView(12, 52, zoom = 6) %>%
addHeatmap(lng= ~Lon, lat = ~Lat, intensity = ~welpen_sum, blur = 20, radius = 15, max = 0.05) %>%
addCircleMarkers(~Lon,~Lat, stroke=FALSE, fillColor = "black",fillOpacity = 1, radius=2, popup = ~as.character(Territorium))